COVID Authorship Community Detection with Machine Learning
Early grad-school experiment: mapped COVID-19 co-authorship to see how different clustering methods reveal the research network. Built a large collaboration graph, compared Infomap clusters to GraphSAGE embeddings, and visualized the differences.
What I explored
- Parsed LitCovid data into a co-author network of ~300K authors and 2M+ edges using Pandas and NetworkX.
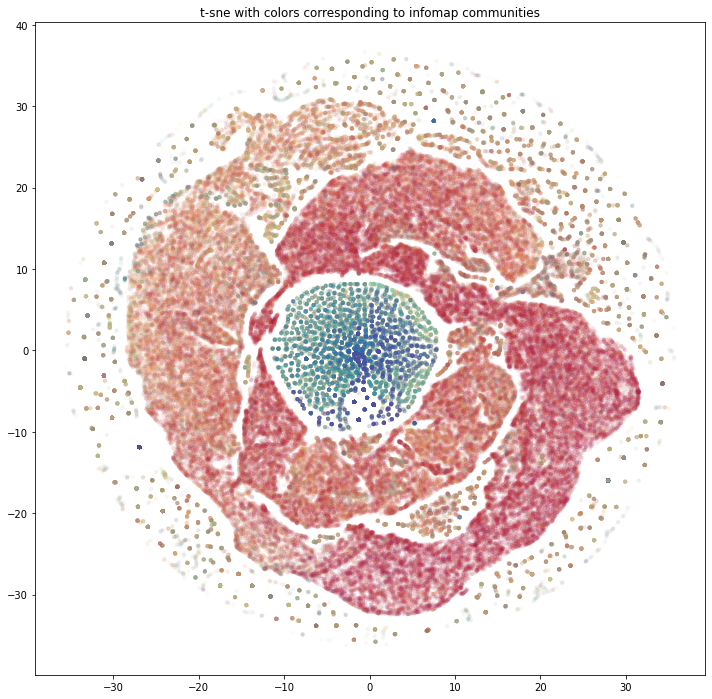
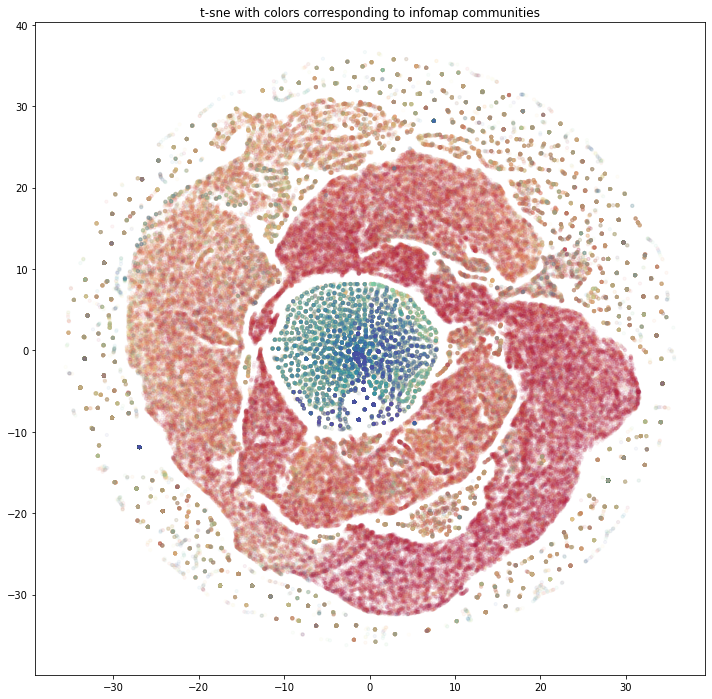
- Ran Infomap for community labels and GraphSAGE embeddings + t-SNE/DBSCAN to see where methods align or diverge.
- Visualized clusters to compare tight algorithmic communities versus looser embedding neighborhoods.
Takeaways
- The two methods surface different shapes of the network—useful reminder to test multiple approaches before interpreting clusters.
- Notebooks keep everything reproducible; see the GitHub repo.